Test kits for residual DNA from host cells
A CHO cell or Escherichia coli is used as a host cell for expression of antibodies or proteins in the manufacturing process of biopharmaceuticals. As DNA of these host cells may cause tumor formation, it is necessary to control the DNA as a process-related impurity.
Guidelines of the World Health Organization (WHO), the Food and Drug Administration (FDA), and the European Pharmacopoeia (EP) state that the final amount of the residual DNA from host cells should be less than 100 pg/dose, or even as little as 10 ng/dose.
FUJIFILM Wako Pure Chemical Corporation offers a DNA extraction and purification kit, “DNA Extractor® Kit”, as a reagent for residual DNA testing. In addition, we have newly released “QCdetect™ Residual DNA Detection Kit for CHO Cells” for detection and quantification of residual DNA from CHO cells.
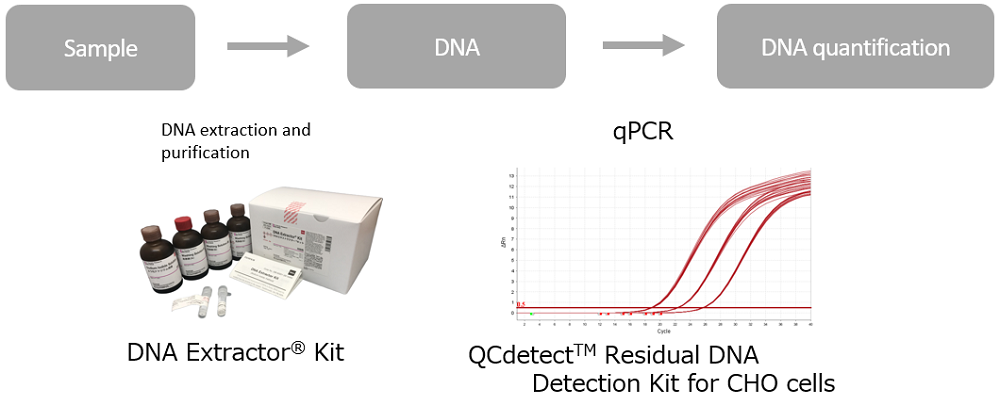
- QCdetect™ Residual DNA Detection Kit for CHO cells
- DNA Extractor® Kit
- Product List
- Related Information
QCdetect™ Residual DNA Detection Kit for CHO cells
Features
A kit for quantifying genomic DNA from CHO cells by qPCR (probe method)
This product is a kit for quantifying genomic DNA from CHO cells by qPCR (probe method). It can be used for residual DNA testing for antibody pharmaceuticals and their manufacturing processes.

- Highly sensitive detection: ≥ 0.0003 pg
- Calibration curve with high linearity
- Minimal inter-assay variability and high reproducibility
- Easy operation using the pre-mix buffer
- Less susceptible to impurities in samples
- Contains an internal control
-
Kit Contents
Product name Quantity 1 x PCR Master Mix 2 x 1 mL DNA Dilution Buffer (DDB) 1 x 10 mL CHO Control DNA 1 x 40 μL -
Detection wavelength
CHO gDNA 520nm (e.g. FAM) Internal Control 554nm (e.g. HEX) Internal Control is non-natural synthetic DNA.
Data
Calibration curve
-
qPCR of 0.0003 pg to 30,000 pg of CHO gDNA was performed using this kit to prepare a calibration curve (n = 3).
A calibration curve with very high linearity was obtained over a wide concentration range from 0.0003 pg to 30,000 pg.

| Pg/well | 0.0003 | 0.003 | 0.03 | 0.3 | 3 | 30 | 300 | 3000 | 30000 |
|---|---|---|---|---|---|---|---|---|---|
| Ct | 37.639 | 34.822 | 31.546 | 28.117 | 24.809 | 21.337 | 18.079 | 14.726 | 11.506 |
| 37.107 | 34.981 | 31.506 | 28.324 | 24.906 | 21.416 | 18.084 | 14.744 | 11.467 | |
| 37.642 | 34.689 | 31.595 | 28.267 | 24.987 | 21.533 | 18.225 | 14.807 | 11.269 | |
| Average Ct | 37.462 | 34.830 | 31.549 | 28.236 | 24.901 | 21.428 | 18.129 | 14.759 | 11.414 |
| S. E | 0.30806 | 0.14652 | 0.04455 | 0.10713 | 0.08904 | 0.09853 | 0.0831 | 0.0424 | 0.12685 |
Detection of fragmented DNA
CHO gDNA was fragmented by sonication and detected by this kit to examine whether the fragmented CHO gDNA could be detected.
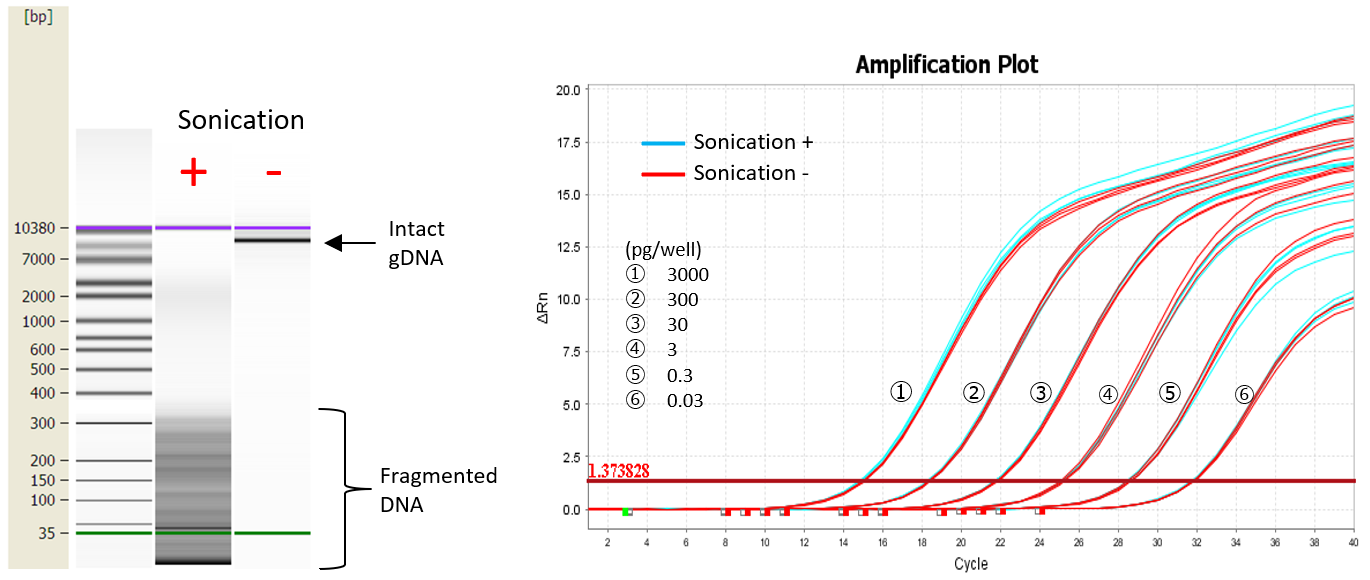
| Fragmented gDNA could be detected with the same sensitivity as intact gDNA. A decrease in detection sensitivity was not observed, even for low concentrations of gDNA. |
Example of use for a high-protein sample -combined with DNA Extractor® Kit-
-
CHO gDNA was spiked to a sample containing high concentration of γ-globulin, and DNA was extracted by DNA Extractor® Kit.
The obtained DNA was quantified with QCdetect™ Residual DNA Detection Kit for CHO cells, and the spike recovery rate was obtained.Sample composition
- 20 mg/mL γ-globulin
- 3% Mannitol
- 2% Sucrose
- 10 mM L-arginine
- 0.01% Tween20
Concentration of spiked CHO gDNA
10 ng/mL
1 ng/mL
100 pg/mL 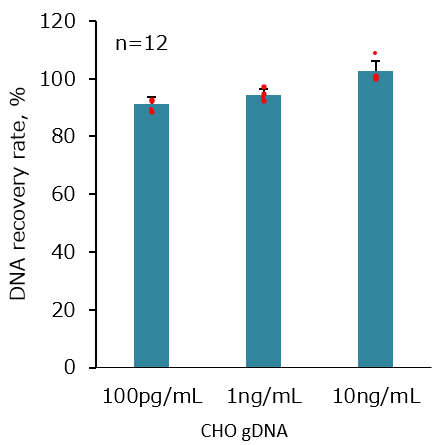
| CHO gDNA could be recovered with a high recovery rate, even when a high-protein sample was used. |
DNA Extractor® Kit
Features
A kit for extracting residual DNA from host cells by sodium iodide method. The extracted DNA can be quantified by qPCR.
This kit can be used for testing and control of the quantity of residual DNA obtained from host cells such as CHO cells, Escherichia coli, yeast, and so on.
- Trace amounts of DNA (100-1,000 fg) can be recovered in high yield.
- No need for tube replacement (all the processes can be completed in the same tube).
- Only 60-90 minutes needed from start to completion of extraction .
- A protocol for high-protein samples is available.
- Sodium iodide method* is adopted.
*The sodium iodide method is a residual DNA extraction technique that is described in the United States Pharmacopeia (USP) 42-NF37, <509> “Residual DNA Testing”.
Kit Contents
| Product name | Quantity |
|---|---|
| Sodium Iodide Solution | 26 mL x 1 |
| Sodium N-Lauryl Sarcosinate Solution | 1.2 mL x 1 |
| Washing Solution (A) | 42 mL x 1 |
| Washing Solution (B) | 40 mL x 2 |
| Glycogen Solution | 0.1 mL x 1 |
Data
Example of spike recovery test
A spike recovery test was performed with gDNA from 3 types of cells: CHO, E.coli, and Pichia pastoris.
CHO gDNA
-
gDNA CHO Spike (fg) Detection (fg) Recovery (%) 100 100 100 1,000 979 98 10,000 10,831 108 100,000 90,031 90 1,000,000 1,045,186 105 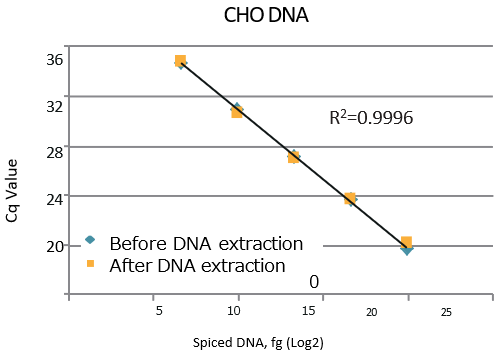
E. coli gDNA
-
gDNA E. coli Spike (fg) Detection (fg) Recovery (%) 1,000 939 94 10,000 11,004 110 100,000 100,009 100 1,000,000 969,911 97 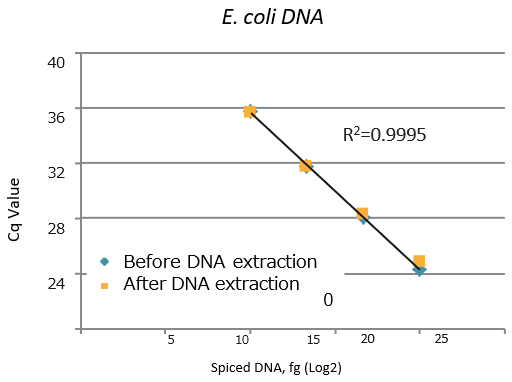
Pichia pastoris GS115 gDNA
-
gDNA Pichia pastoris GS115 Spike (fg) Detection (fg) Recovery (%) 10,000 10,316 103 100,000 94,045 94 1,000,000 1,031,630 103 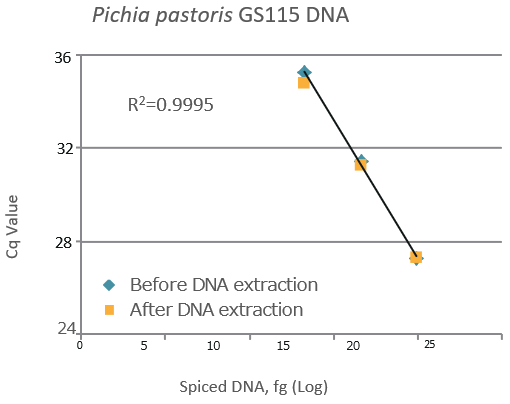
| A high DNA recovery was observed under all conditions. High linearity was observed between the amount of spiked DNA and Cq value. |
Spike recovery test of CHO gDNA using culture supernatant
The spike recovery rate of CHO-derived DNA was determined using culture supernatant.
| Methods: CHO-derived DNA (10 fg to 1 ng) was spiked to culture supernatant of PANC-1 cells and the DNA was extracted with this kit. qPCR of the extracted DNA was performed and Cq value was measured. Separately, qPCR was carried out for purified water spiked with CHO-derived DNA without DNA extraction, and Cq value was measured. This was used as a standard condition. A calibration curve was prepared under the standard condition, and DNA recovery rate was calculated. Sample: 1. Standard condition: purified water spiked with CHO-derived DNA (without DNA extraction): 2. Culture supernatant condition: DNA extracted from PANC-1 cell culture supernatant spiked with CHO-derived DNA using this kit |
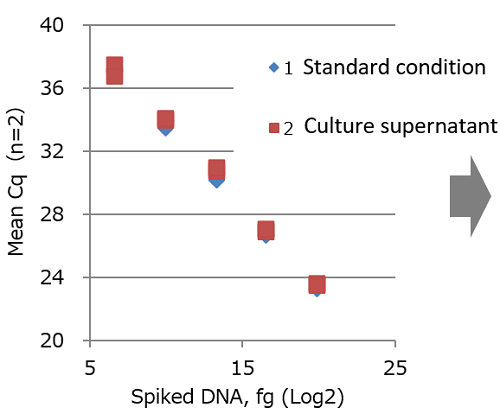
Amount of spiked DNA and mean Cq
1. Standard condition: R2 = 0.9998
2. Culture supernatant: R2 = 0.9982-
Culture supernatant Spiked amount (fg) Volume recovered (fg) Recovery rate (%) 0 ND 10 100 93 93 1,000 741 74 10,000 6,333 63 100,000 86,703 87 1,000,000 874,502 87 A calibration curve was prepared using mean Cq values obtained under the standard condition, and DNA recovery rate was calculated from the mean Cq value obtained under the culture supernatant condition.
| DNA in culture supernatant could be recovered in high yield within the range from 100 fg to 1 ng of spiked DNA. |
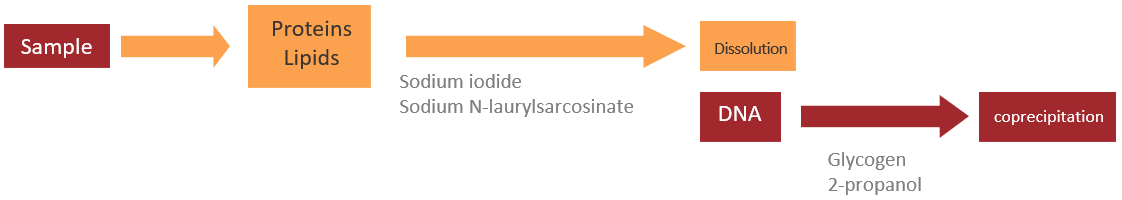
Principle
- Sodium iodide, a chaotropic ion, and sodium N-laurylsarcosinate are added to a sample to solubilize proteins and lipids.
- Glycogen and then 2-propanol are added to the sample, and DNA is coprecipitated with glycogen.
- A DNA pellet is collected.
Product List
- Open All
- Close All
QCdetect™ Residual DNA Detection Kit for CHO cells
DNA Extractor® Kit
Related Information
Category
- Cell Culture
- Biopharmaceuticals Manufacturing
- Quality Control Test
Content
- Quantitative detection of residual DNA with DNA Extractor® KitWako Blog
- Since many biopharmaceuticals, including vaccines, are manufactured from cultured cells or microorganisms such as Escherichia coli, it is pointed out that host cell-derived DNA may remain in resultant…
Product content may differ from the actual image due to minor specification changes etc.